CODA-TB DREAM
We participated in a DREAM challenge to use audio recordings of cough to diagnose the presence of Tuberculosos in a patient.
Authors
Gautam Ahuja[1], Aakash Rao[1], Diya Khurdiya[1], Shalini Balodi[1], Ashwin Salampuriya[1], Rintu Kutum[1,2,*]
Affiliations:
- Department of Computer Science, Ashoka University, Haryana, India
- Trivedi School of Biosciences, Ashoka University, Haryana, India
*. Corresponding Author
Description
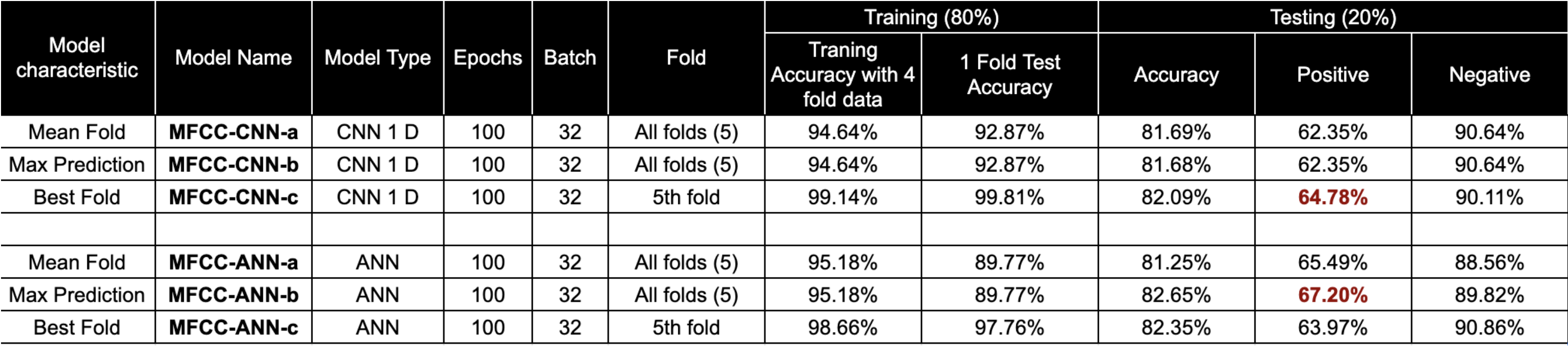
MFCC based neural network classifier to predict TB status from cough sound. We achieved best class-wise accuracy of 67.20 % for positive with mel-frequency cepstral coefficients (MFCC)[cite] as features and artificial neural network (ANN) model.
- The audio file of a TB patient differentiates from a non-TB patient to a human ear. There is a bit of “heaviness” in a positive cough audio. The relative pitch is lower, however, this inherited differentiation is hard for a machine to replicate.
- To tackle this, two approaches were selected: mel-frequency cepstral (MFCC) coefficients and scaled log2 mel-frequency spectrogram points. The “Mel” (or melody) approach provides a tool to compare pitches.
- The MFCC(s) of audio can be used to uniquely identify the audio. The aim is to train a model such that it picks up on these “unique” features to differentiate a TB/non-TB patient.
- The MFCC(s) of audio are extracted and linearised twice using mean (40 features) and standard deviation (40 features). 80 features were used for modeling
- For scaled log2 mel-frequency spectrogram, we linearised the log2 mel-frequency spectrogram (2816 features) and later values were scaled in between 0 to 1
Methods
MFCC:
Utilizing the mfcc() function from the librosa library available in python [1], features were extracted for each audio file. Next, the 40x22 feature matrix underwent two linearizations. Once to 40x1 using a mean on columns. And then to a 40x1 by applying a standard deviation to the columns a second time. We used 80 features per audio file to build the models (ANN & CNN 1D)
Scaled log2 mel-frequency spectrogram:
For the scaled log2 mel-frequency spectrogram, we extracted the spectrogram points for each audio file using feature.melspectrogram() function from the python librosa library. Then a log2 transformation was performed on the obtained points. We linearised the log2 mel-frequency spectrogram (2816 features for each file) and later values were scaled in between 0 to 1.
Training & testing
The data was split into an 80:20 ratio to generate a training set (7817 files) and testing set (1955 files), respectively. During training phase, a 5-fold split was performed on the training data using the KFold() function from the sklearn library. The model was then trained five times, validating on each fold while training on the other four in each iteration. Finally, testing was performed in 20% test set.
ANN based model:
This is a sequential model with three dense layers and a softmax layer. The first dense layer has 100 neurons, followed by a second layer that has 200 neurons and a drop of 0.5. The third layer also has 100 neurons. An activation function of RELU was used on all the above layers. The final layer is a dense layer with a softmax layer for binary classification. The model was compiled using a categorical cross-entropy loss function and an Adam optimizer.
CNN 1D-based models:
This is a sequential model with three Conv1D() layers, one flatten(), one dense layer, and one softmax layer. The first layer has 32 filters and a kernel size of 2. The second layer has 64 folders and a kernel size of 2. And the third layer has 128 filters and a kernel size of 2. The flatten layer is used to reduce the dimension and then fed into a dense layer with 128 neurons. All layers used an activation function of RELU. The final layer is a dense layer with a softmax layer for binary classification. The model was compiled using a categorical cross-entropy loss function, SGD optimizer, and a learning rate of 0.01.
Results:
Code repository
Github repository: https://github.com/rintukutum/dream-coda-tb-sc1-chsxashoka
My Role
I adviced on the project in terms of the design of the Artificial Neural Network as well as the benchmarking of other algorithms on the dataset.
Acknowledgements
We would like to acknowledge Mphasis F1 foundation, Centre for Computation Biology & Bioinformatics (BIC) computing facility & High Performance Computing (HPC) at Ashoka University for the support.
License
Please refer to the CODA-TB Licensing guidelines